Walrus is redefining the scientific process by transforming experimental data into a secure, decentralized commons that ensures longevity, accessibility, and reproducibility. Researchers deposit datasets—from genomic sequences and particle physics events to climate simulations—into versioned blob repositories, where cryptographically signed provenance guarantees that every result can be traced back to its origin. By capturing instrument calibration logs, environmental conditions, and analysis scripts alongside raw data, Walrus allows experiments to be rerun exactly as originally conducted, tackling the reproducibility challenges that affect a majority of published biomedical and physical science findings.
Capturing Complete Experimental Lineage
Instruments connect seamlessly to Walrus through vendor-neutral adapters: electron microscopes stream high-resolution TIFF stacks annotated with lens metadata, mass spectrometers log ion fragmentation patterns with precise voltage readings, and telescopes embed atmospheric conditions within FITS headers. Automated capture preserves every nuance, prioritizing critical signal data even under bandwidth constraints. Metadata schemas align with cross-disciplinary standards, such as Genomics Standards Consortium for sequencing and IUCR dictionaries for crystallography, enabling discovery across research domains while maintaining domain-specific fidelity.
Versioned blobs allow granular referencing—individual gel lanes, specific collision events, or single sequencing reads—so researchers can cite exact experimental units rather than entire datasets. Incremental encoding captures only changes between successive runs, minimizing storage overhead without compromising reconstruction capabilities. Interactive lineage graphs visualize transformations, linking raw signals through analysis pipelines to final results, enabling traceable, clickable provenance back to every measured parameter.
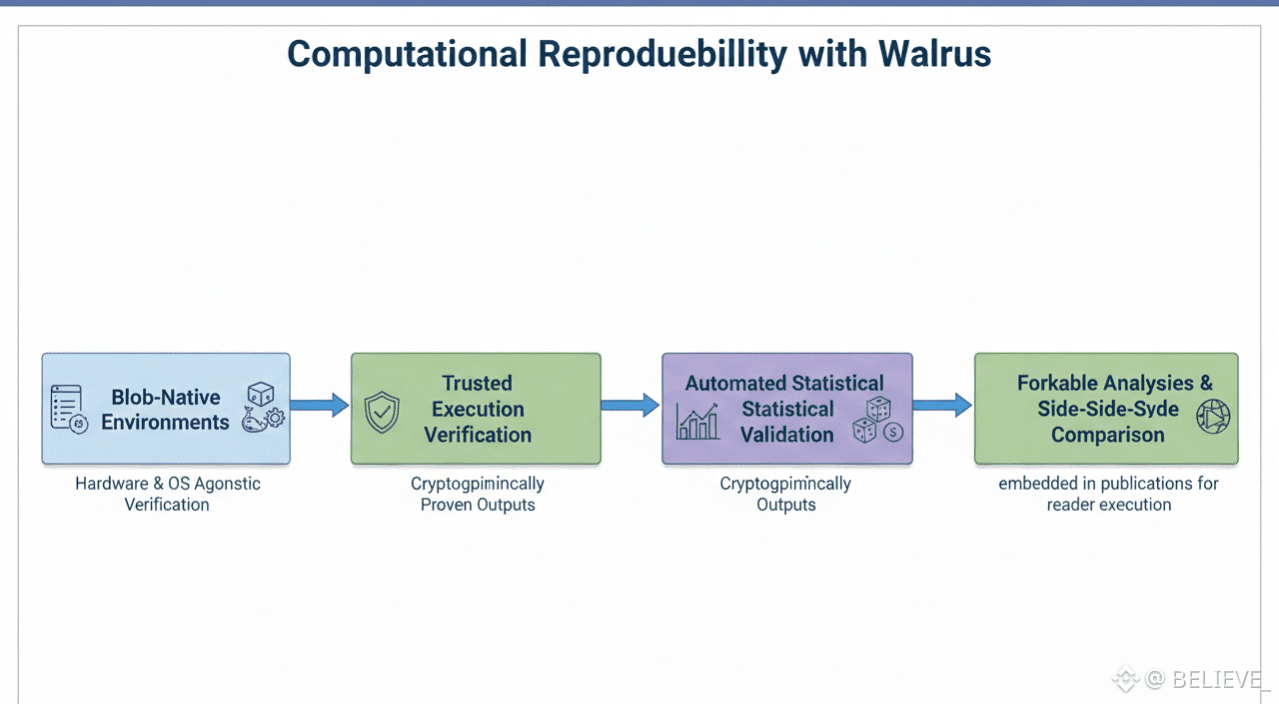
Ensuring Computational Reproducibility
Walrus packages full computational environments as self-contained blob compositions, including Jupyter notebooks with fixed library versions, Docker containers with precise runtime dependencies, and workflow managers preserving scheduler states. Trusted execution environments verify reproducibility, generating cryptographic attestations that numerical outputs remain identical across hardware generations and operating systems. Researchers can fork analyses and compare side-by-side outcomes without exposing proprietary code.
Automated statistical validation engines rerun analyses against original parameters, flagging anomalies and potential biases. Monte Carlo simulations, random seeds, and stochastic models are fully traceable, ensuring identical distributions decades after initial runs. Publications can embed live reconstruction links, allowing readers to execute exact computations directly from figure legends rather than static summaries.
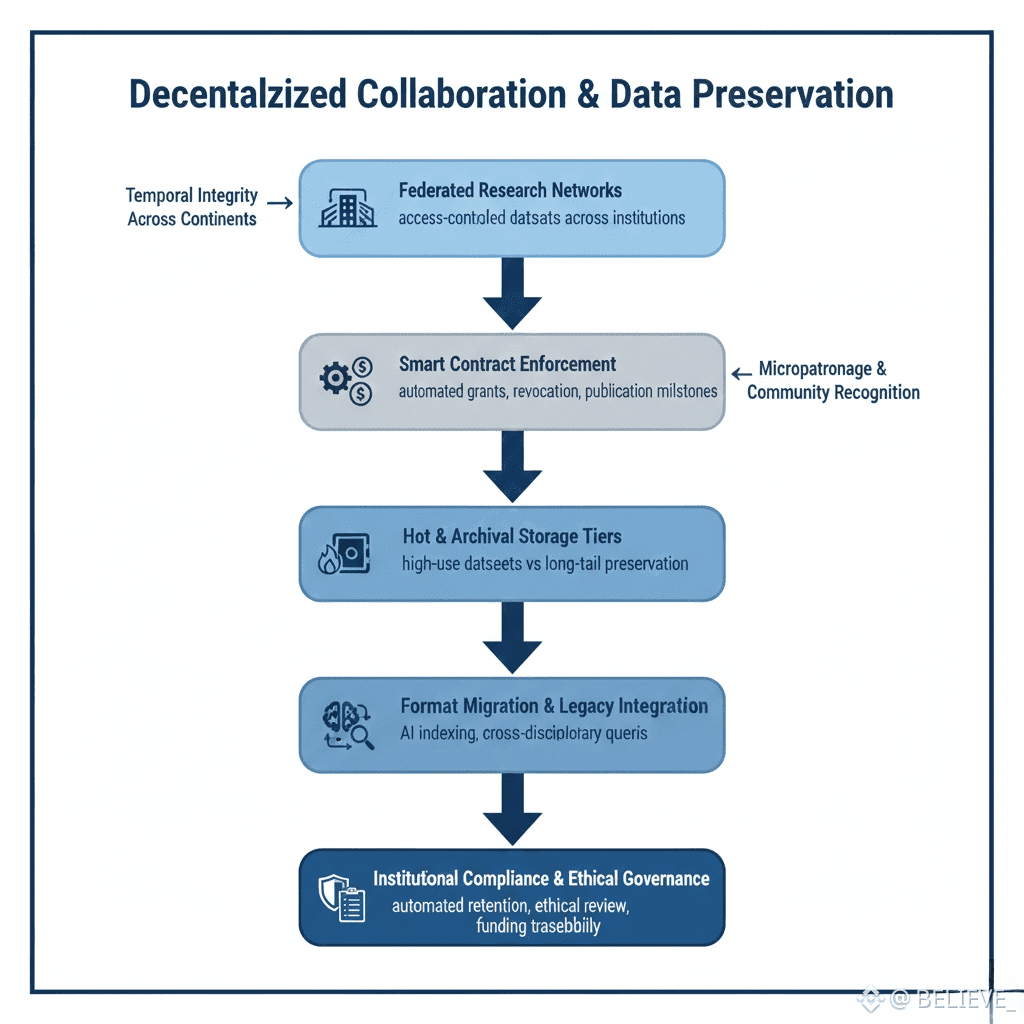 Cross-Institutional Collaboration at Scale
Cross-Institutional Collaboration at ScaleWalrus enables federated research networks with fine-grained access control. NIH-funded datasets may be restricted to US-based investigators for a defined exclusivity period, while ERC or global physics data mandates immediate open sharing. Multi-institution consortia synchronize through federated blob namespaces, maintaining local administrative control while publishing unified aggregates for meta-analysis. Smart contracts enforce data use agreements, automatically revoking or granting access based on grant cycles or publication milestones.
Real-time collaboration surfaces live instrument streams to remote co-authors, with lag-compensated visualization ensuring temporal integrity across continents. Shared computational resources pool HPC allocations, dynamically routing analyses while preserving experimental ownership. Concurrent modifications are resolved through timestamped reconciliation, creating alternative version histories that preserve every dataset variant for future verification.
Preserving Rare and Long-Tail Data
Walrus applies intelligent storage tiers based on anticipated reuse. High-impact datasets—such as novel protein structures—reside in hot storage for millisecond access, while specialized or niche datasets cascade to archival tiers with guaranteed reconstruction windows. Format migration engines automatically convert legacy file types, like early Affymetrix CEL files or VAX FORTRAN outputs, into self-describing containers without data loss.
Grant lifecycle integration tracks storage against budgets, generating NSF-compliant data management plans. Community micropatronage sustains orphaned datasets, allowing researchers citing preserved work to contribute micro-fees proportionally. Recognition programs highlight datasets of exceptional reproducibility and reuse potential, establishing academic incentives parallel to conventional publications.
Advanced Analytics and Knowledge Discovery
Walrus enables blob-native analysis across disciplines. Genomic researchers can perform federated GWAS without centralizing samples, climate scientists query petabyte-scale reanalysis datasets, and astronomers correlate multi-wavelength surveys spanning decades. AI-powered indexing accelerates search over unstructured data, revealing hidden correlations and suggesting testable hypotheses grounded in statistical rigor.
Visualization platforms progressively render complex datasets, from electron density maps to multi-exposure telescope imagery. Collaborative whiteboards link analyses across labs, enabling real-time interactive exploration where insights in one dataset dynamically filter related data elsewhere. Publication-quality figures embed reconstruction pipelines, ensuring reviewers can independently reproduce every panel.
Institutional Integration and Compliance
Walrus bridges enterprise and university repositories with bidirectional sync adapters, preserving decentralized permanence while reflecting institutional catalogs. Library systems index blob inventories alongside journal publications via OAI-PMH feeds. Funding agencies monitor ROI by tracing research contributions from deposit to citation and commercial translation, and tenure committees can evaluate impact through verifiable dataset usage metrics. Automated compliance scanning flags lapses in retention policies, enforcing data management requirements with minimal manual oversight.
Community Stewardship and Ethical Governance
Walrus fosters sustainable scientific culture through community-guided preservation. Advisory councils allocate resources to endangered or underrepresented datasets, while peer-review processes extend to data itself, assessing completeness, documentation, and reuse potential. Ethical review boards protect human subjects, balancing transparency with privacy and de-identification standards. Disciplinary working groups harmonize metadata across institutions, creating globally interoperable research archives. Career recognition incentivizes exemplary data stewardship alongside high-impact publications.
Walrus transforms experimental outputs from ephemeral project artifacts into resilient, reproducible, and communal knowledge infrastructure. Complete provenance ensures every finding can be retraced. Computational environments guarantee repeatability. Collaboration frameworks span institutions seamlessly. Long-tail preservation safeguards rare discoveries. Advanced analytics unlock cross-disciplinary insights. Institutional integration enforces compliance. Ethical governance sustains trust. Researchers gain a platform where scientific progress compounds over generations, building knowledge as an interconnected web rather than isolated papers, bridging disciplines and historical epochs.


